pharokka
pharokka is a fast phage annotation pipeline.

phold
If you like pharokka, you will probably love phold. phold uses structural homology to improve phage annotation. Benchmarking is ongoing but phold strongly outperforms pharokka in terms of annotation, particularly for less characterised phages such as those from metagenomic datasets.
pharokka still has features phold lacks for now (identifying tRNA, tmRNA, CRISPR repeats, and INPHARED taxonomy search), so it it recommended to run phold after running pharokka.
phold takes the Genbank output of Pharokka as input. Therefore, if you have already annotated your phage(s) with Pharokka, you can easily update the annotation with more functional predictions with phold.
Google Colab Notebooks
If you don't want to install pharokka or phold locally, you can run pharokka and phold, or only pharokka, without any code using the Google Colab notebook https://colab.research.google.com/github/gbouras13/pharokka/blob/master/run_pharokka_and_phold.ipynb
Overview
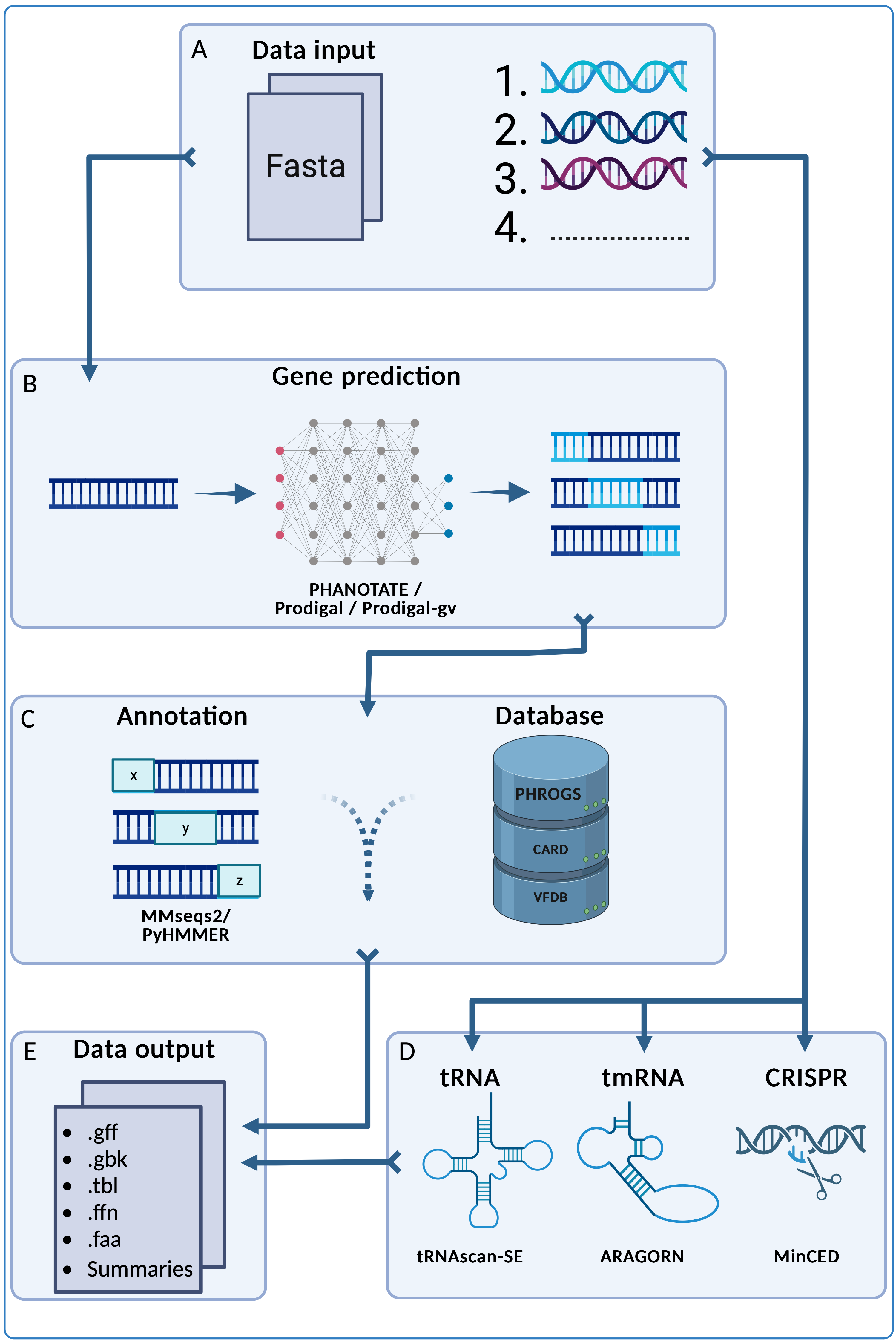
pharokka uses PHANOTATE, the only gene prediction program tailored to bacteriophages, as the default program for gene prediction. Prodigal implemented with pyrodigal and Prodigal-gv implemented with pyrodigal-gv are also available as alternatives. Following this, functional annotations are assigned by matching each predicted coding sequence (CDS) to the PHROGs, CARD and VFDB databases using MMseqs2. As of v1.4.0, pharokka will also match each CDS to the PHROGs database using more sensitive Hidden Markov Models using PyHMMER. Pharokka's main output is a GFF file suitable for using in downstream pangenomic pipelines like Roary. pharokka also generates a cds_functions.tsv file, which includes counts of CDSs, tRNAs, tmRNAs, CRISPRs and functions assigned to CDSs according to the PHROGs database. See the full usage and check out the full documentation for more details.
Manuscript
For more information, please read the pharokka manuscript:
George Bouras, Roshan Nepal, Ghais Houtak, Alkis James Psaltis, Peter-John Wormald, Sarah Vreugde, Pharokka: a fast scalable bacteriophage annotation tool, Bioinformatics, Volume 39, Issue 1, January 2023, btac776, https://doi.org/10.1093/bioinformatics/btac776